Mozyme setpi charge=-2 Di-anion of hydroquinone C 0.00000000 0 0.0000000 0 0.0000000 0 C 1.42891759 1 0.0000000 0 0.0000000 0 1 0 0 C 1.39123760 1 121.7152363 1 0.0000000 0 2 1 0 C 1.42884933 1 121.6862338 1 0.0522686 1 3 2 1 C 1.42731168 1 116.6123504 1 -0.0326433 1 4 3 2 C 1.39298952 1 121.7002715 1 0.0291866 1 5 4 3 O 1.28854675 1 121.6283321 1 179.9905509 1 1 2 3 O 1.28863750 1 121.5839611 1 179.9914815 1 4 3 2 H 1.07825816 1 117.6731944 1 -179.9642741 1 2 1 3 H 1.07822008 1 120.6215632 1 -179.9762574 1 3 2 1 H 1.07805743 1 117.7610380 1 -179.9848142 1 5 4 3 H 1.07809519 1 120.5409971 1 179.9757231 1 6 5 4 2 3 1 6 4 5
If the geometry is supplied using GEO_DAT, then SETPI cannot be used. Instead use SETPI=<file>.TXT, for example, setpi=setpi_for_Cytochrome-P450.txt or setpi="../setpi for Cytochrome-P450.txt. In this example, the keyword points to a file in the next-higher level. The file pointed to by setpi should contain exactly the same data as defined earlier.
When large biomolecules are being run, defining atoms by their atom number is sometimes difficult. In a PDB atom label, the number may or may not be the same as the atom number. Two simpler methods for defining atoms are provided: the PDB label and the JSmol label. If the PDB or JSmol label is used, then enclose the label in quotation marks. Wildcards, where the chain letter is replaced by an asterisk, or a residue is replaced by three asterisks, are allowed. The first occurrence that matches will be used. Examples of valid labels are:
| Examples of the use of PDB and JSmol definitions of atoms used in π-bonds |
|
| "[8OG]1157:A.C6" | "[8OG]1157:A.N1" |
| "CB PHE B 74" | "CG PHE B 74" |
| "[8OG]1157:A.C6" | "N1 8OG B1157" |
| The same examples, with wildcards | |
| "[***]1157:A.C6" | "[***]1157:*.N1" |
| "CB PHE * 74" | "CG PHE * 74" |
| "[8OG]1157:*.C6" | "N1 *** B1157" |
| Quotation marks are part of the definition | |
There is no need to supply all the π bonds, only
those necessary to resolve any ambiguities. Other examples of the need
for SETPI include:
Distinguishing the anion of p-hydroxy N-methyl pyridine from its quinone type
tautomer.
Ensuring that meso-tetra(para-N-methylpyridinato)-porphyrin
has the correct charge.
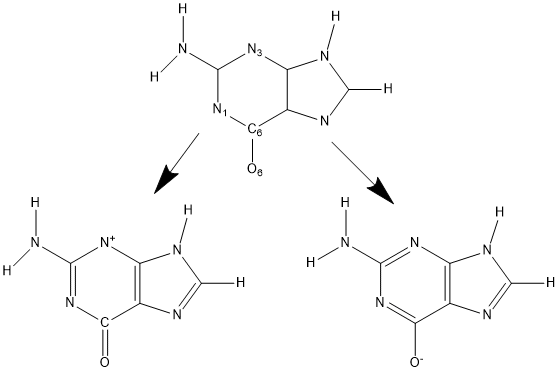
Guanine (see top figure) anion can be made by removing a proton from either O6 or N1. If the only guide to the electronic structure is the topology, then both of the lower figures are valid. If a double bond is made between C6 and O6 (bottom left figure) then N3 would have only two valencies used, instead of the three valencies expected. This is clearly wrong but it follows logically from the presence of the C=O double-bond. In MOPAC, by default, a nitrogen atom with a valence other than 3 would be assigned a positive charge. This is obviously the wrong charge. To prevent this from happening, SETPI could be used to define a double bond between C6 and N1. If that is done (bottom right figure) then all the faults are corrected. C6 now automatically forms a single bond with O6, which then automatically gets a negative charge, the ring becomes aromatic, and the faulty divalent nitrogen becomes trivalent.
See also CVB to explicitly make or break bonds, and atom labels to explicitly assign charges.
Other examples of the use of SETPI in defining the Lewis structure can be seen in the following ARC files::
1-Myoglobin (1M6C) This protein contains a heme ring system, a particularly complicated Lewis structure. CVB is used to break chemical bonds to isolate the porphorin ring from the iron atom, then SETPI is used to lock the π-system so that the charge on the ring is correct.
Human MTH1 (3ZR0) More π-bonds than necessary were used in defining the Lewis structure for the benzimidazole. Only one is really necessary, but extra π-bonds don't cause problems.
Metalloprotein (1J3F) For details of why this was a difficult system, see Notes on Metalloprotein